About MangoPanBase
MangoPanBase is an academic research resource focused on mango pangenome construction. It hosts data, analysis results, and interactive visualization tools to support the scientific community.
🧬 Our Pangenome Construction
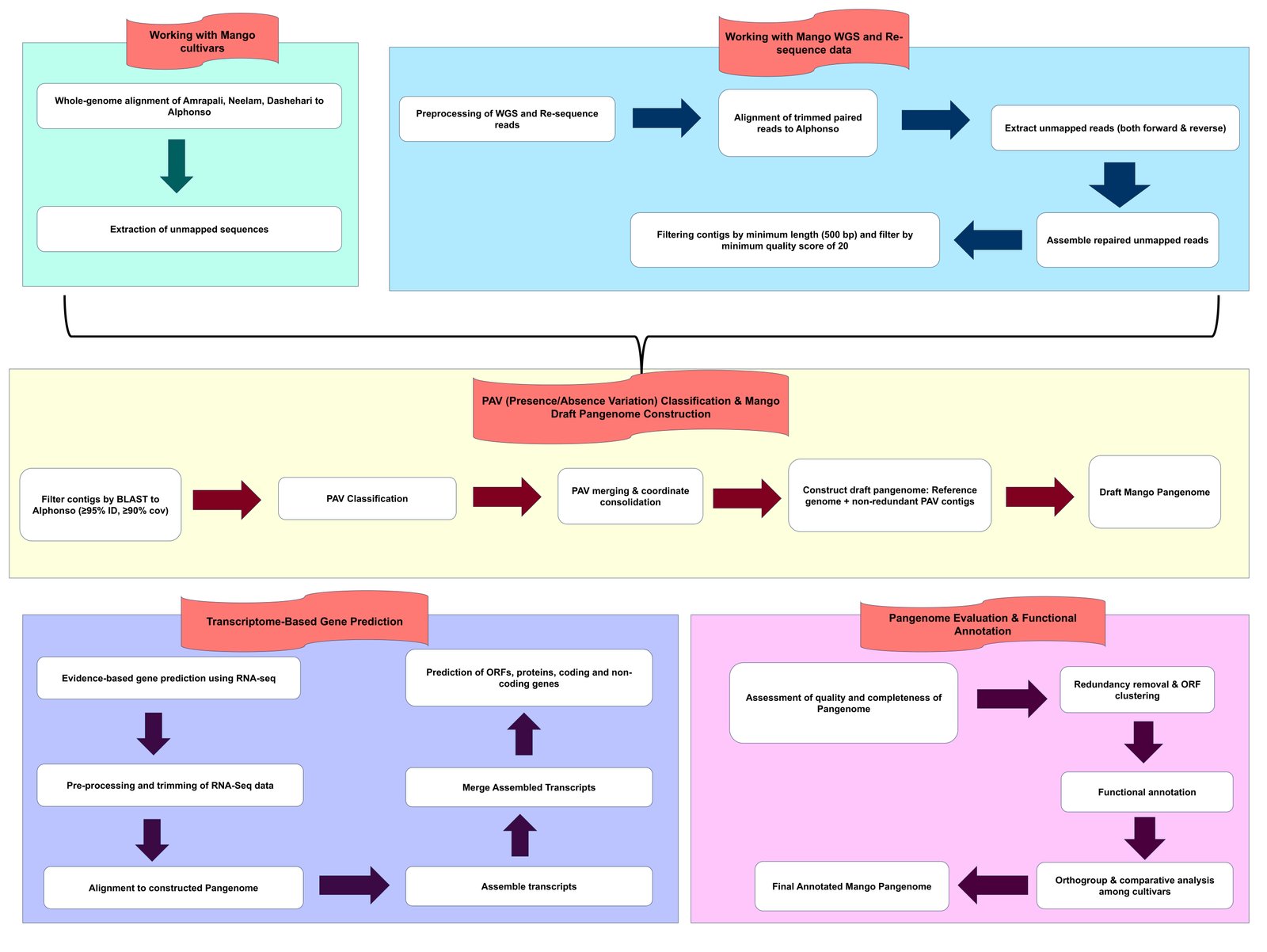
Constructed using 4 mango cultivars: Alphonso, Amrapali, Neelam, and Dashehari and 36 WGS and resequenced datasets.
Combined sequence-based assembly (488.95 Mb) .
Achieved a high-quality assembly with N50 of 17.65 Mb.
📊 Key Results
- 50,090 predicted proteins in the non-redundant protein set
- 13,111 orthogroups identified, with 3,375 core orthogroups shared across all cultivars
- BUSCO completeness: 96.5%, reflecting high assembly accuracy
- 11,416 significant GO terms and 3,923 KEGG pathways mapped
-
Unique gene counts per cultivar:
- Alphonso (2,588)
- Amrapali (2,239)
- Dashehari (725)
- Neelam (1,071)
🌐 Pangenome Browser
The Interactive Mango Pangenome Browser enables genome exploration with integrated tools:
- Visualizes gene distribution, orthogroups, and annotation results across cultivars
- Supports comparative analysis of core, dispensable, and unique genes
- Provides integrated functional annotations (GO, KEGG) for downstream research
🔗 Analysis Workflow Overview
This pipeline illustrates the construction and analysis steps of the Mango Pangenome.
Contact
For questions, please contact the MangoPanBase team.
